Environmental monitoring of pharmaceutical cleanrooms provides assurance that our production environment is in control, and in compliance. A variety of test methods are performed to monitor the number of total particles in the air, the number of viable (alive) particles in the air and the number of viable organisms on surfaces, including personnel working in the controlled rooms.
The number of particles in the air can be determined very quickly. Particle counters are used to collect a known quantity of air and then print out the breakdown of the particle sizes and the number of each size particle in that sample. Results print out within minutes. This allows for quick reactions, and allows areas with high particle counts to be shut down, investigated and their condition corrected during the production of a batch. In addition, drug compounds produced while areas are out of specification can be segregated.
Unfortunately, these results do not tell us when we have viable organisms in the environment. Methods for testing for viable organisms require that the organisms be collected, either with an air sampler or with swabs and/or contact plates. The sampling media is then incubated, typically for three to seven days, and then colonies are counted. This delay to results delays reaction to contamination issues and can make investigations very difficult. By that time, the rooms in question have typically been cleaned numerous times, so re-sampling results are almost always meaningless and determining the root cause of the contamination is difficult. Since real-time response is not possible, batches are jeopardized.
However, rapid microbiological methods may provide a solution. For some time, the industry has been interested in using RMM for sterility testing, to expedite product release. While a desirable goal, it comes with a great deal of additional regulatory scrutiny. Some companies have been successful in implementing RMM for release testing in the U.S., but acceptance of rapid methods for finished product testing is still not widespread and regulatory opinions on the topic vary. At the very least, a regulatory filing is needed to change from the traditional sterility test to an alternative method.
As rapid microbiological methods evolve, their use to perform environmental monitoring could provide a path of less resistance from a regulatory standpoint. Many of the technologies that have been developed for product testing may have significant merit for performing environmental monitoring (Table 1). It remains to be seen whether there will be a significant market for this kind of service (Box, bottom).
| Company | System | Technology; Uses |
| bioMerieux | BacT/ALERT | Detects CO2 from growing organisms; used for qualitative product testing and analysis of swab samples |
| BioVigilant | IMD-A 200-1, IMD-A 220-4 | Optical technology; instantaneous detection of particles and microorganisms in air samples |
| Celsis | RapiScreen, AMPiScreen | ATP bioluminescence; qualitative product testing and analysis of swab samples |
| Chemunex | Chem Scan RDI | Solid-phase cytometry; quantitative testing of water, filterable products |
| Lonza | microCompass | Real-time reverse transcriptase PCR assay; quantitative testing of water, filterable products, swab samples |
| Millipore | Milliflex Quantum | Fluorescent staining based system; water, filterable products, swab samples |
| Pall | Pallchek | ATP bioluminescence; quantitative of water, filterable product testing |
| Rapid Micro Biosystems | Growth Direct System | Automated non-destructive growth-based system; quantitative testing of water, filterable products, swab samples |
Case Study: Use of a Rapid Method to Analyze Swab Samples
A study was performed at the MicroWorks laboratory in Crown Point, Indiana, to determine if it might be feasible to utilize an alternative technology to analyze swab samples. Two different test methodologies were performed.
Experimental Background
The purpose of surface sampling as part of an overall environmental monitoring program is to track the level of surface contamination in the facility to ensure that cleaning and sanitization is effective. Swabs are often used for sampling irregular or hard-to-reach surfaces and critical surfaces where contact plates are not practical. In addition, cleaning hold time studies are often performed using swabs. Sanitizers collected from surfaces can be neutralized and dilutions can be done when highly contaminated areas are sampled.
Surface samples are most frequently incubated at 30-35˚C for at least three days or 20-25˚C for five to seven days, or at some combination of the two temperatures.
For this study, we used the MicroWorks Swab Sampling System (MSSS 001). The swab system consists of a sterile, double-bagged, gamma-irradiated product containing a calcium alginate swab and a sodium citrate diluent. These sterile packages are designed to be taken into cleanrooms and/or isolator systems.
The instrument utilized for this study was the BacT/ALERT system. This automated microbial detection system is built on bioMerieux’s colorimetric technology that detects microorganism growth by tracking CO2 production. To utilize this system, sterile liquid media bottles are inoculated with the test sample and the bottle is loaded into the instrument. A variety of different media are available depending on the product being tested and the target organisms. If microorganisms are present in the test sample, carbon dioxide is produced as the microorganisms metabolize the substrates in the culture medium. With CO2 production, the color of the sensor in the bottom of each culture bottle changes from dark to light.
A light-emitting diode (LED) projects light onto the sensor every ten minutes. The light reflected is measured by a photodetector. As more CO2 is generated, more light is reflected. This information is compared to the initial sensor reading. If there is a high initial CO2 content, an unusually high rate of CO2 production, and/or a sustained production of CO2, the sample is determined to be positive. If the CO2 level does not change significantly after a specified number of days at optimal conditions, the sample is determined to be negative.
Immediately upon detection, positive results are indicated and by an audible beep. If no microbial growth is present after a specified time, a sample is determined to be negative.
Our hypothesis was that, using this methodology, the swab samples could be analyzed and test results generated faster than they could by way of more traditional methods.
Experiment 1
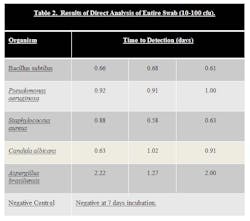
Stainless Steel coupons were inoculated with ATCC strains of B. subtilus, C. albicans, A. brasiliensis, P. aeruginosa, and S. aureus at a level of 10-100 cfu. Surfaces were then swabbed and the samples loaded into the system.
The results can be seen in Table 2.
Experiment 2
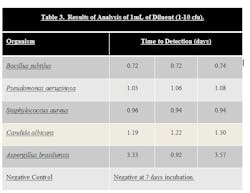
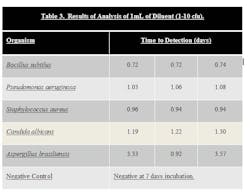
Stainless steel surfaces were inoculated with ATCC strains of B. subtilus, C. albicans, A. brasiliensis, P. aeruginosa, and S. aureus at a level of 10–100 cfu. Surfaces were swabbed and the swab was added to the 10 ml sodium citrate diluent that is provided in the test kit. The tube of sodium citrate containing the swab was then vortexed to dissolve the swab. One mL of the sodium citrate diluent was then added to the media bottle using a syringe, resulting in an inoculum level of 1-10 cfu. The bottle was then loaded into system. Results can be seen in Table 3.
Experimental Observations and Conclusions
Actual analysis of the swab samples took a matter of minutes. Loading the samples took about five minutes per sample. Since the test system alarms when a positive sample is detected, there was a fast response time. Only negative samples needed to continue to incubate for the duration of the incubation period. There was no subjectivity of the test results as seen with conventional methods. Since swabs could be placed directly into the bottles, there was less chance of contaminating the sample.
Based on the results, it appears that this RMM system is a feasible alternative for analysis of swab samples for surface contamination. Low levels of organisms (1-10 cfu) can be detected utilizing this methodology.
Samples that were inoculated with bacterial cultures at the 10-100 inoculum level were determined to be positive in 0.58 to 1.00 days. Samples inoculated with yeast and mold at the 10-100 inoculum level were determined to be positive in 0.63 to 2.22 days. In both cases, this is a significant time savings compared to traditional incubation times.
Samples that were inoculated with bacterial cultures at the 1-10 inoculum level were determined to be positive in 0.72 to 1.08 days. Samples inoculated with yeast and mold at the 1-10 inoculum level were determined to be positive in 1.22 to 3.57 days.
Granted, this was a small experiment and only a few organisms were tested. The results were encouraging, but of course future experimentation will be needed to further support this methodology.
The Future of RMM for Environmental Monitoring
As presented in Table 1, there are many choices for rapid technologies, and many of these technologies could potentially be utilized for environmental monitoring tests. The case study presented here is only one of many possibilities. Some technologies may be able to give us answers in hours rather than days. Faster results will ultimately mean environmental monitoring will be more meaningful.
As these technologies emerge, industry will need to weigh the pros and cons of implementing them. The big advantage, of course, is a faster result, which means more control and safer products. The biggest drawback currently is cost. Reagents, test systems and disposables needed for rapid technologies will add upfront cost to the environmental monitoring program. In addition, there is an initial cost for the systems and the validation efforts. These pros and cons will need to be assessed and balanced, but clearly the technology continues to show great promise.
|
Will RMM’s Make Their Mark in EM? In that environmental monitoring for surfaces requires testing surface swabs or air filters, various RMM technologies could be used. Incubation, sample prep, and testing using the Celsis RMM system can usually be done in less than a day, notes Lori Daane, Ph.D., a microbiologist and the company’s VP. Still, environmental monitoring is usually a “value-add” or a “nice to have,” Daane says, but not the main reason that manufacturers are looking for a rapid microbiological system. The volume for EM is typically low, and if there are contaminants they will typically need an accurate CFU count, which is usually done via traditional methods. Rather, the next step for environmental monitoring is to understand a facility’s persistent organisms at the genetic level, Daane says. Strain typing enables a lab to know if, for example, a particular strain of S. aureus is the same one that’s been appearing in their products or a new one recently introduced (or mutated, for that matter). |
About the Author
Dawn McIver is founder and President of MicroWorks, Inc., which provides testing, consulting and training services to the pharmaceutical and medical device industry. Dawn has a Bachelor of Science degree in Biology from Purdue University and over twenty years experience in Microbiology. Her first six years in the field included positions of Research Microbiologist and Microbiology Laboratory Manager for Silliker Laboratories. She then moved into the pharmaceutical industry, working for Fujisawa, USA as the Microbiology Laboratory Supervisor. Dawn has authored chapters in “Laboratory Validation, A Practitioner’s Guide” and “Environmental Monitoring, Volume 1” as well as a chapter in the just-published “Environmental Monitoring, Volume 3”.